Meta-analysis of cytotoxic T lymphocyte antigen-4 (CTLA-4) genetic polymorphisms and Graves’ disease
Abstract
Background: Graves’ disease (GD) is an autoimmune disorder specific to the thyroid, characterized by a complex etiology influenced by both genetic and environmental factors. The CTLA-4 gene, known for its involvement in immune regulation, has been scrutinized regarding its potential contribution to GD susceptibility, particularly concerning the rs3087243 polymorphism. We conducted a meta-analysis to explore the association between this genetic variation and the likelihood of GD development.
Methods: We thoroughly searched the PubMed, Medline, and EMBASE databases, as well as the reference lists of pertinent articles published up to 2024. Studies examining the association between GD and the CTLA-4 CT60 polymorphism were selected for inclusion. Data extraction and statistical analyses were conducted using Review Manager 5.4 software, which assessed multiple genetic models and the publication bias.
Results: Six studies encompassing 1904 controls and 926 GD cases met the inclusion criteria. Our meta-analysis uncovered a substantial correlation between the CTLA-4 (rs3087243) polymorphism and GD across multiple genetic models (allele, homozygous, dominant, and recessive), suggesting the potential role of this genetic variant in predisposing individuals to GD. An examination of publication bias revealed symmetrical funnel plots.
Conclusion: Our meta-analysis provides evidence for a significant association between GD and the CTLA-4 (rs3087243) polymorphism, highlighting its potential contribution to GD susceptibility. Additional investigation utilizing larger sample sizes and diverse populations is essential to corroborate these findings and clarify the exact mechanisms governing the association between the CTLA-4 gene and GD.
Citation
Kalarani I B, Veerabathiran R. Meta-analysis of cytotoxic T lymphocyte antigen-4 (CTLA-4) genetic polymorphisms and Graves’ disease. Eur J Transl Clin Med. 2025;8(2):35-42Introduction
Autoimmune diseases (ADs) encompass more than 100 conditions distinguished by the disrupted regulation of inflammatory mechanisms targeting one or more autoantigens [1]. Autoimmune thyroid disease (AITD) is the most prevalent form of organ-specific AD [2]. AITDs, which impact over 5% of the population, are the most common autoimmune diseases. These include Graves’ disease (GD) and Hashimoto’s thyroiditis (HT). Additionally, abnormal thyroid function varies across populations, from 1-2% in males to 7-9% in females [3]. The development of autoantibodies and inflammation result in hypothyroidism. On the other hand, GD is distinguished by the generation of antibodies against the thyroid-stimulating hormone receptor (TSHR), which leads to overstimulation and excessive release of thyroid hormone by thyrocytes. Thyroid-specific genes are the genetic components implicated in etiopathogenesis [4].
GD impacts approximately 0.5% of the global population, yet its occurrence has not been explicitly documented within the Indian population [5]. While environmental factors (e.g. infection, stress) play a significant role in triggering GD in susceptible individuals, research involving twins has indicated that genetic factors are the primary influencers, responsible for approximately 80% of the predisposition to GD [6]. Additionally, both linkage and association studies have identified the involvement of genes related to thyroid function and genes responsible for immune regulation in GD pathogenesis [7]. The immune-regulatory genes associated with GD development are also linked to other autoimmune disorders. These genes include CTLA-4, HLA-DR, PTPN22, CD40, thyroglobulin, and the TSH receptor [8]. A co-stimulatory molecule that promotes peripheral self-tolerance and inhibits T-cell activation is encoded by the CTLA-4 gene, which is found on chromosome 2q33. Autoimmune disorders have been associated with modifications in CTLA-4, including the functional polymorphism CT60 (rs30807243). The G allele of CT60 is associated with the reduced expression of soluble CTLA-4 (sCTLA-4) compared to the A allele [9]. When CTLA-4 is dysregulated or genetically altered, it affects the T cell’s capacity to inhibit immunological activation, which can result in autoimmunity, including GD [10]. The aim of this study was to evaluate the correlation between the +6230 G/A (rs3087243) single nucleotide polymorphisms (SNPs) and GD using the meta-analysis methodology.
Material and methods
A comprehensive literature search was conducted to identify relevant studies for inclusion in the meta-analysis. To minimize bias and ensure the thoroughness of the review, both authors independently screened articles based on pre-established inclusion and exclusion criteria. Any discrepancies between reviewers were resolved through discussion. Data from the selected studies were extracted in duplicate, with any inconsistencies addressed in the same manner, further ensuring the accuracy and reliability of the results.
Literature search
We searched the literature published up to 2024 using PubMed, Medline and EMBASE. The search terms “Graves’ disease”, “GD”, and “cytotoxic T lymphocyte-associated antigen 4”, “CTLA-4”, combined with “polymorphism” were used. All research investigating the relationship between GD and the CTLA-4 (rs3087243) polymorphism was carefully chosen and thoroughly examined. In the interim, additional articles were identified by reviewing the references of the studies.
Inclusion and exclusion criteria
All studies that were published in English were taken into account for this meta-analysis and had to fulfill the subsequent inclusion requirements: case-control studies, genotype distribution or allelic frequency data were available for comparison, sufficient data were available to calculate an OR along with a 95% CI. The following were among the exclusion criteria: studies with overlapping data, studies where family members were examined (linkage considerations were the basis for the analysis) and studies where the control population’s genotype distribution was outside the Hardy-Weinberg equilibrium (HWE). A PRISMA flowchart illustrating the inclusion and exclusion of studies is provided in Figure 1.
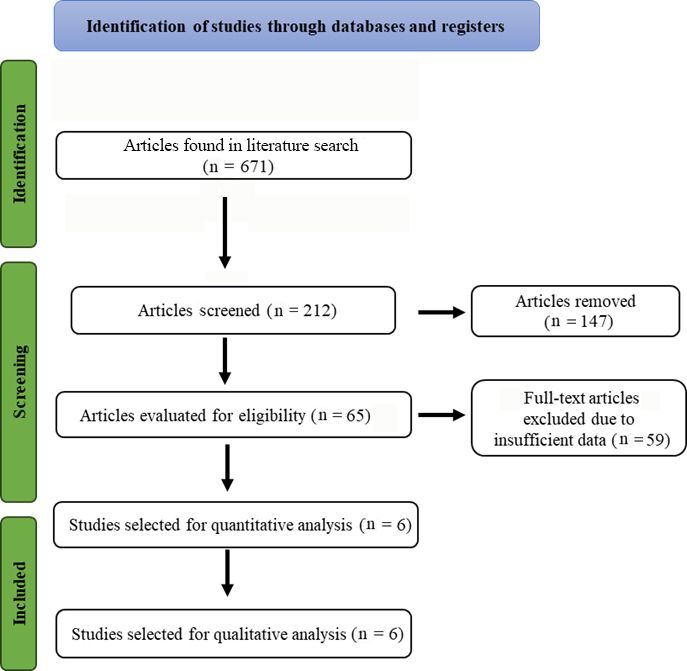
Figure 1. Study selection of the CTLA-4 gene polymorphism and Graves’ disease
Data extraction
The data collected from each study include information such as the title, the primary author, the year of publication, the place of origin, the study design and research methods, the genotyping strategy used and the specific goals of the study. We did not address or account for potential discrepancies or conflicting results among the articles included in the analysis.
Quality assessment
The Newcastle-Ottawa Scale (NOS) was employed to assess the quality of case-control studies [11]. Using this tool we evaluated each study’s comparability, outcomes, and methodological integrity, including sampling techniques, response rates, and representativeness. Studies scoring 7 out of 10 were considered suitable, a threshold determined after reviewing relevant literature-based meta-analyses.
Statistical analysis
We used the Review Manager 5.4 software (The Cochrane Collaboration, London, UK) for data analysis, with the statistical significance set at p < 0.05 for all genetic variants. Essential tools and procedures were utilized to conduct comprehensive genetic association meta-analyses, assess the clinical relevance of genetic variations, ensure significance testing in large-scale genetic studies, and maintain statistical power. The heterogeneity assumption from previous research was evaluated using the Chi-square-based Q statistic test and the I2 metric value. Significant data were identified with a p-value of less than 0.1. The preceding research used the random effect model to assess the odds ratio, accompanied by a 95% CI. An analysis of the HWE was conducted using the Chi-square test [12]. A forest plot, incorporating 95% confidence intervals and the overall odds ratio, was generated to evaluate the strength of the association between gene polymorphism and autoimmune thyroid disease. Publication bias in the meta-analysis was assessed through a funnel plot.
Power analysis
Using the GPower 3.1 tool (The G*Power Team, Heinrich-Heine-Universität Düsseldorf, Germany), we computed the statistical power under a 95% confidence interval with an α error probability set at 0.05 [13]. For each selected gene, the power of the case and control study sample sizes was combined and analyzed separately.
CTLA-4 gene interaction network analysis
To better understand the role of CTLA-4 and its potential functional partners in GD, we used the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) online server (STRING Consortium, http://string-db.org/) to build a network of CTLA-4 gene interactions [14].
Results
Of the 671 studies on the correlation between the collected data and GD, 6 specifically investigated the relationship with the CTLA-4 (rs3087243) polymorphism and contained data from 926 GD cases and 1904 controls. These articles were then examined to see which ones fulfilled the inclusion criteria for our study and had valuable data. Detailed information regarding each study we included is outlined in Table 1, along with the characteristics of the cases and controls Figure 1. Study selection of the CTLA-4 gene polymorphism and Graves’ disease regarding the correlation between GD and the CTLA-4 polymorphism. These 6 articles included patients from diverse ethnic groups [9, 15-19].
Table 1. Characteristics of studies included in the meta-analysis
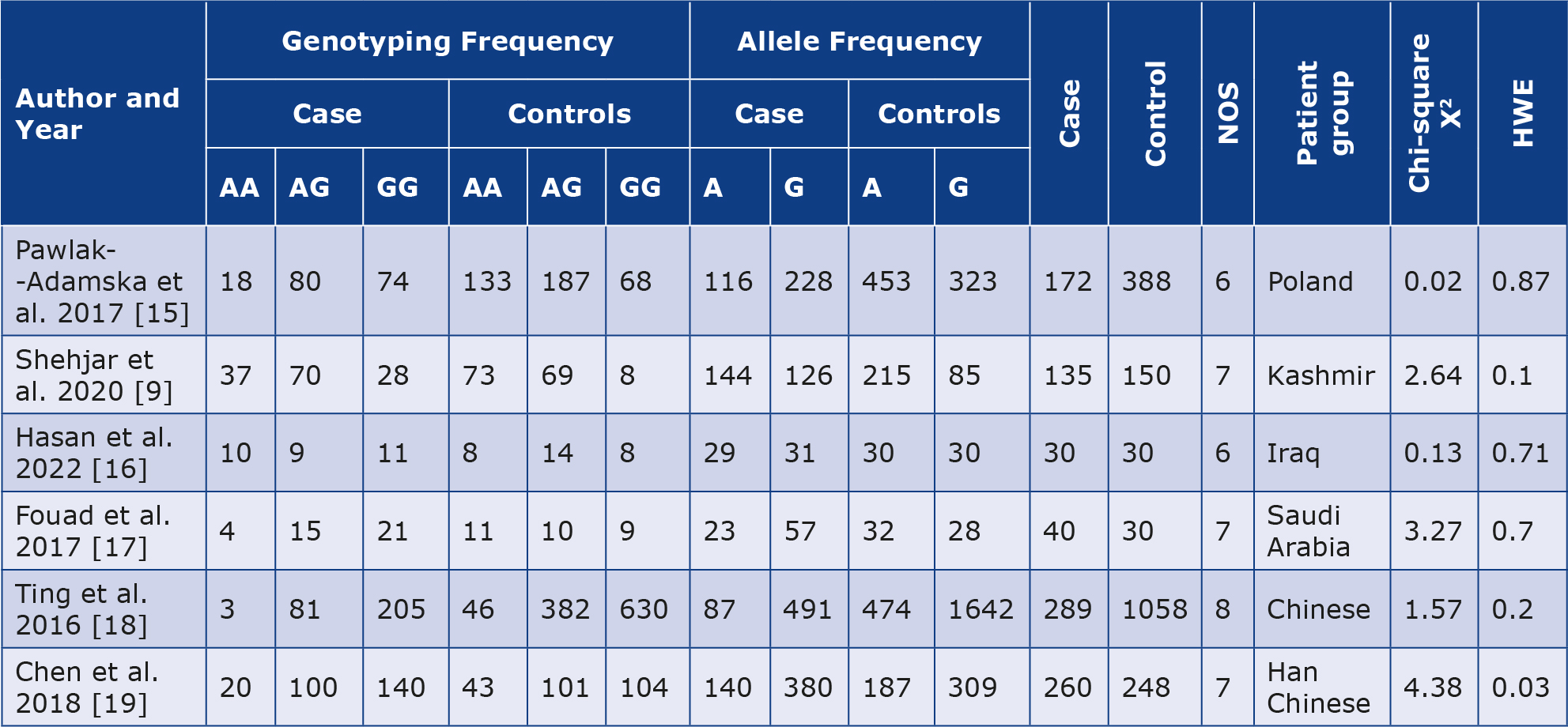
NOS – Newcastle-Ottawa Scale; HWE – Hardy-Weinberg Equilibrium
Publication bias and the analysis of quantitative data
We conducted 6 investigations to assess the relationship between AITD, particularly GD, and the CTLA-4 gene (rs3087243) polymorphism. Our research revealed a strong correlation between GD risk and CTLA-4 polymorphism in various genetic models. The allele model (A vs. G) exhibits an I2 of 65%, an OR of 1.96, a 95% CI of 1.53- 2.50, and a p-value < 0.00001. In the homozygous model (AA vs. GG), the I2 is 56%, presenting an OR of 4.48, with a 95% CI of 2.57-7.82 and a p-value of 0.0001. The dominant model (AA+AG vs GG) showcases an OR of 2.83, a 95% CI of 1.82–4.40, and a p-value of 0.0001. Meanwhile, the recessive model (AA vs. AG+GG) displays an I2 of 68%, an OR of 2.29, a 95% CI of 1.57–3.34, and a p-value of less than 0.0001 under the random effects model. Fixed effects were preferred when the I2 was 39% in the heterozygote model (AG vs. GG), yielding an OR of 0.58, with a 95% CI of 0.48–0.70 and a p-value of less than 0.0001 (Figure 2). We employed a funnel plot (Figure 3) and Egger’s regression test in our investigation to assess publication bias. The funnel plot suggests that the dataset may contain small-study effects or publication bias. The majority of research indicates a positive correlation between GD and CTLA-4 polymorphisms. We used the 3DSNP tool to provide additional information on the relationship between chromosomes and SNPs [20]. As a result, we created Circos plots, which offer an intricate and precise representation of the whole genomic dataset displayed in Figure 4. The genomic landscape of the CTLA-4 gene area on chromosome 2q33 is highlighted by this Circos plot (Figure 4), which displays important SNPs (like rs3087243), gene-gene closeness (like CD28, ICOS), and potential linkage disequilibrium or functional connections. With internal arcs indicating strong genetic interactions that may affect CTLA-4 expression and function, important to autoimmune disorders like GD disease, the clustered red SNPs indicate locations of high polymorphism.
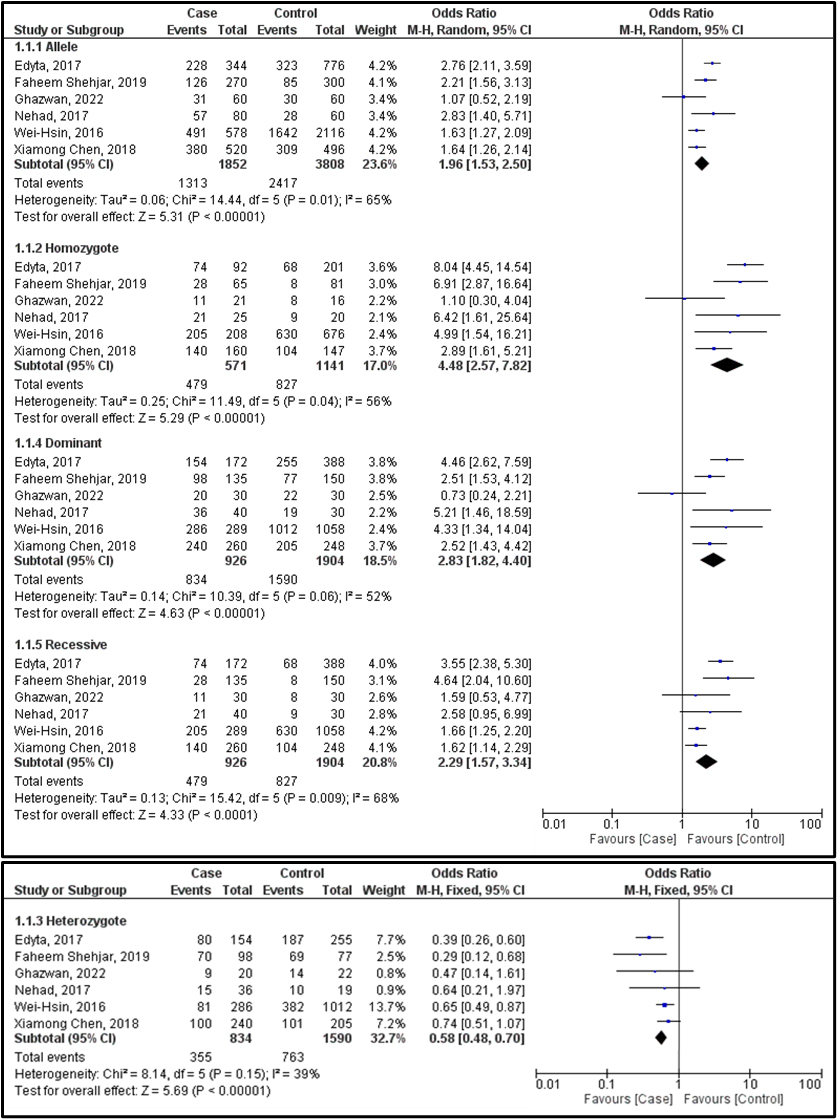
Figure 2. Forest plot showing the association between the CTLA-4 gene polymorphism and Graves’ disease in all 5 models
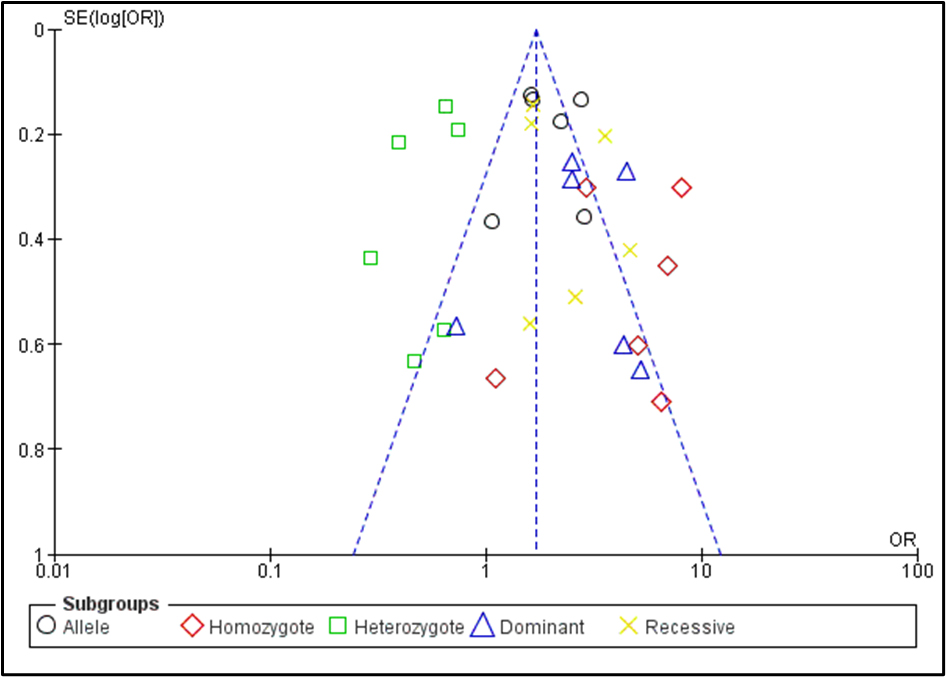
Figure 3. Publication bias in the association between the CTLA-4 gene polymorphism and Graves’ disease in all models
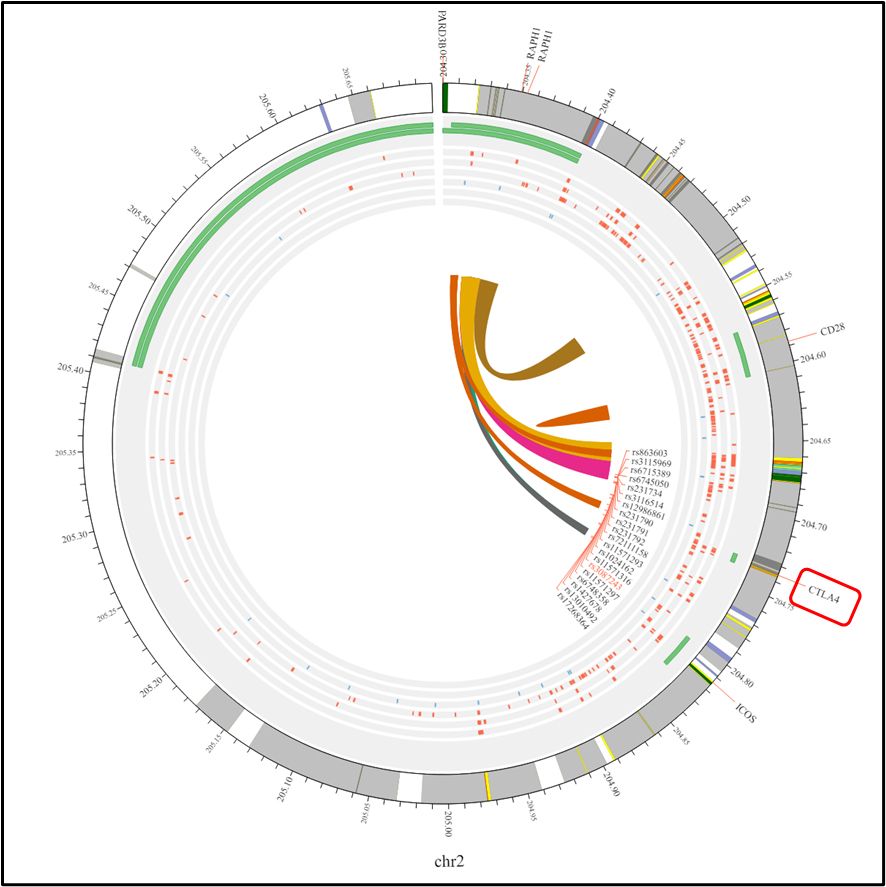
Figure 4. Circos plot illustrating the chromosomal interactions among rs3087243 polymorphisms
Power analysis
We conducted a power analysis to determine the significance level for each selected SNP. Our results indicated that the sample sizes from the selected literature were sufficient to meet this stringent significance level, ensuring the robustness and reliability of our findings. This comprehensive approach confirms that our meta-analysis can detect significant associations for the SNPs under investigation, as shown in Figure 5.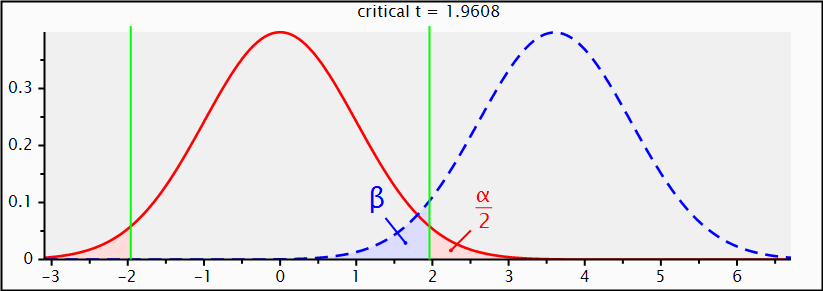
Figure 5. Power analysis result of the CTLA-4 gene
Gene-gene network and interactions
Our STRING online server analysis indicated that CTLA-4 interacts with several genes. The 10 most significant genes from the network of gene-gene interactions are shown in Figure 6. According to these results, CTLA-4 exhibits high-confidence functional connections with other important immune regulators, such as PD-1, PD-L1, CD28, ICOS, and galectin-9 (LGALS9). Because of these connections, CTLA-4 is positioned as a key participant in immune homeostasis maintenance, and as a crucial target in the modulation of autoimmune diseases.
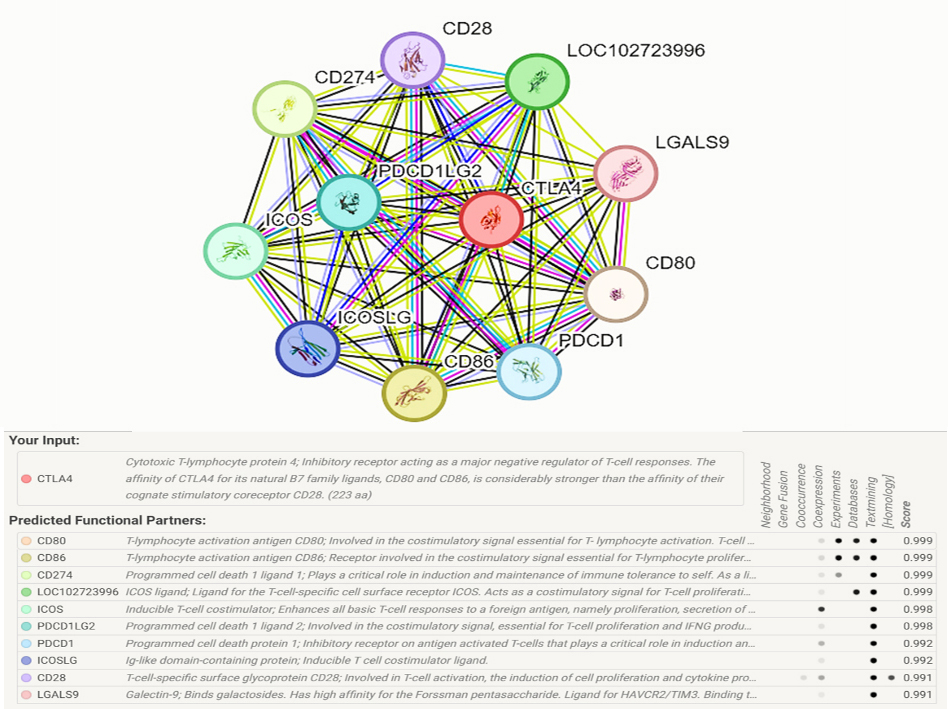
Figure 6. Human CTLA-4 interaction network with other genes obtained from the STRING server
Discussion
This meta-analysis, which comprised 1904 controls and 926 GD patients, investigated the relationship between the CTLA-4 (rs3087243) polymorphism and GD. Egger’s test results offered statistical support for the symmetry of the funnel plot. According to the overall findings, all genetic models showed a strong (p < 0.01) association between variant genotypes and GD risk. Although GD has severe clinical significance, its pathomechanism is still unclear. However, stress and infection play a significant role in the susceptibility of specific individuals to GD. SNPs help to analyze genetic changes and the likelihood of developing certain diseases. GD is a thyroid-specific auto immune disease mediated by antibodies [21]. The CTLA-4 protein is susceptible to autoimmunity and can send an inhibitory signal to T-cells [22]. Si et al. identified the CTLA-4 protein as the gatekeeper of conjugation timing; decreased conjugation may offer protection against extended durations of time that cytotoxic T-cells spend close to autoantigen-defined targets [22]. That finding has received much attention because of its significant contribution to autoimmunity [22]. Our meta-analysis showed a strong (p < 0.01) correlation between the CTLA-4 polymorphism and GD. In addition, we report that there may be a relationship between the risk of GD and the CTLA-4 (rs3087243) polymorphism. In conclusion, our meta-analysis demonstrates a strong correlation between increased susceptibility to GD illness and the CTLA-4 (rs3087243) polymorphism, specifically the G allele. These findings emphasize the genetic role of CTLA-4 in GD and the necessity for further investigation into its functional significance and potential as a therapeutic target or biomarker.
While our study provides valuable insights, it has several limitations that need to be considered. First, our sample size was relatively small, which may limit the generalizability of the findings and reduce the statistical power, particularly in subgroup analyses. Additionally, the potential for population stratification could have influenced the results, as genetic variations may differ across populations. Although the funnel plots appeared symmetrical (suggesting no significant publication bias), we acknowledge that this visual inspection method has limitations and a more formal test for publication bias could provide further clarity. Furthermore, due to the small number of studies included, we were unable to perform a meta-regression or a more in-depth exploration of certain sources of heterogeneity. Given these limitations, further research with larger sample sizes and more diverse populations is needed to confirm and expand upon the observed associations between the CTLA-4 gene and GD.
Conclusion
Our meta-analysis investigated the relationship between GD risk and the CTLA-4 (rs3087243) polymorphism, involving 1904 controls and 926 GD patients across six studies. In several genetic models, our research revealed a strong correlation between the CTLA-4 polymorphism and GD, indicating that this genetic variation may play a role in predisposing people to GD. The CTLA-4 gene, critical in regulating T-cell activity, has garnered attention for its involvement in autoimmune diseases, including GD. Despite the strengths of our analysis (e.g. comprehensive literature search and robust statistical methods), it has limitations, including the small sample sizes in some of the included studies. Well-designed research is warranted to elucidate the precise relationship between the CTLA-4 gene and GD susceptibility.
Conflict of interest
None.
Funding
None.
References
| 1. |
Bieber K, Hundt JE, Yu X, Ehlers M, Petersen F, Karsten CM, et al. Autoimmune pre-disease. Autoimmun Rev [Internet]. 2023;22(2):103236. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1568997222002063.
|
| 2. |
McLeod DSA, Cooper DS. The incidence and prevalence of thyroid autoimmunity. Endocrine [Internet]. 2012;42(2):252–65. Available from: http://link.springer.com/10.1007/s12020-012-9703-2.
|
| 3. |
Miteva MZ, Nonchev BI, Orbetzova MM, Stoencheva SD. Vitamin D and Autoimmune Thyroid Diseases - a Review. Folia Med (Plovdiv) [Internet]. 2020;62(2):223–9. Available from: https://foliamedica.bg/article/47794/.
|
| 4. |
Ramgopal S, Rathika C, Padma MR, Murali V, Arun K, Kamaludeen MN, et al. Interaction of HLA-DRB1* alleles and CTLA4 (+ 49 AG) gene polymorphism in Autoimmune Thyroid Disease. Gene [Internet]. 2018;642:430–8. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0378111917310223.
|
| 5. |
Weetman AP. Graves’ Disease. N Engl J Med [Internet]. 2000;343(17):1236–48. Available from: http://www.nejm.org/doi/abs/10.1056/NEJM200010263431707.
|
| 6. |
Tomer Y. Mechanisms of Autoimmune Thyroid Diseases: From Genetics to Epigenetics. Annu Rev Pathol Mech Dis [Internet]. 2014;9(1):147–56. Available from: https://www.annualreviews.org/doi/10.1146/annurev-pathol-012513-104713.
|
| 7. |
Eschler DC, Hasham A, Tomer Y. Cutting Edge: The Etiology of Autoimmune Thyroid Diseases. Clin Rev Allergy Immunol [Internet]. 2011;41(2):190–7. Available from: http://link.springer.com/10.1007/s12016-010-8245-8.
|
| 8. |
Shehjar F, Afroze D, Misgar RA, Malik SA, Laway BA. Association of polymorphic variants of IL-1β and IL-1RN genes in the development of Graves’ disease in Kashmiri population (North India). Hum Immunol [Internet]. 2018;79(4):228–32. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0198885918300442.
|
| 9. |
Shehjar F, Dil-Afroze, Misgar RA, Malik SA, Laway BA. A significant association of the CTLA4 gene variants with the risk of autoimmune Graves’ disease in ethnic Kashmiri population. Cell Immunol [Internet]. 2020;347:103995. Available from: https://linkinghub.elsevier.com/retrieve/pii/S000887491930293X.
|
| 10. |
Simone R, Tenca C, Fais F, Luciani M, De Rossi G, Pesce G, et al. A Soluble Form of CTLA-4 Is Present in Paediatric Patients with Acute Lymphoblastic Leukaemia and Correlates with CD1d+ Expression. Moormann AM, editor. PLoS One [Internet]. 2012;7(9):e44654. Available from: https://dx.plos.org/10.1371/journal.pone.0044654.
|
| 11. |
Wells GA, Shea B, O’Connell D, Peterson J, Welch V, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses [Internet]. The Ottawa Hospital. Research Institute. Department of Epidemiology and Community Medicine, University of Ottawa; 2000 [cited 2025 Aug 1]. Available from: https://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.
|
| 12. |
Rodriguez S, Gaunt TR, Day INM. Hardy-Weinberg Equilibrium Testing of Biological Ascertainment for Mendelian Randomization Studies. Am J Epidemiol [Internet]. 2009;169(4):505–14. Available from: https://academic.oup.com/aje/article-lookup/doi/10.1093/aje/kwn359.
|
| 13. |
Faul F, Erdfelder E, Buchner A, Lang A-G. Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behav Res Methods [Internet]. 2009;41(4):1149–60. Available from: http://link.springer.com/10.3758/BRM.41.4.1149.
|
| 14. |
Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res [Internet]. 2023;51(D1):D638–46. Available from: https://academic.oup.com/nar/article/51/D1/D638/6825349.
|
| 15. |
Pawlak-Adamska E, Frydecka I, Bolanowski M, Tomkiewicz A, Jonkisz A, Karabon L, et al. CD28/CTLA-4/ICOS haplotypes confers susceptibility to Graves’ disease and modulates clinical phenotype of disease. Endocrine [Internet]. 2017;55(1):186–99. Available from: http://link.springer.com/10.1007/s12020-016-1096-1.
|
| 16. |
Hasan GA, Altamemi IA. CTLA-4 Polymorphism along with Proinflammatory Cytokines in Autoimmune Thyroiditis Disease. Wiadomości Lek [Internet]. 2022;75(3):577–83. Available from: https://wiadlek.pl/wp-content/uploads/archive/2022/03/WLek202203103.pdf.
|
| 17. |
Fouad NA, Saeed AM, Mahedy AW. Association of CTLA-4+ 49 A/G and CT60 gene polymorphism with Graves’ disease. Egypt J Immunol [Internet]. 2017;24(2):63–70. Available from: https://www.bu.edu.eg/portal/uploads/Medicine/INTERNAL MEDICINE/297/publications/Ahmed Wageh Mahdey _3.pdf.
|
| 18. |
Ting W-H, Chien M-N, Lo F-S, Wang C-H, Huang C-Y, Lin C-L, et al. Association of Cytotoxic T-Lymphocyte-Associated Protein 4 (CTLA4) Gene Polymorphisms with Autoimmune Thyroid Disease in Children and Adults: Case-Control Study. Pietropaolo M, editor. PLoS One [Internet]. 2016;11(4):e0154394. Available from: https://dx.plos.org/10.1371/journal.pone.0154394.
|
| 19. |
Chen X, Hu Z, Liu M, Li H, Liang C, Li W, et al. Correlation between CTLA-4 and CD40 gene polymorphisms and their interaction in graves’ disease in a Chinese Han population. BMC Med Genet [Internet]. 2018;19(1):171. Available from: https://bmcmedgenet.biomedcentral.com/articles/10.1186/s12881-018-0665-y.
|
| 20. |
Lu Y, Quan C, Chen H, Bo X, Zhang C. 3DSNP: a database for linking human noncoding SNPs to their three-dimensional interacting genes. Nucleic Acids Res [Internet]. 2017;45(D1):D643–9. Available from: https://academic.oup.com/nar/article-lookup/doi/10.1093/nar/gkw1022.
|
| 21. |
Pastuszak-Lewandoska D, Sewerynek E, Domańska D, Gładyś A, Skrzypczak R, Brzeziańska E. CTLA-4 gene polymorphisms and their influence on predisposition to autoimmune thyroid diseases (Graves’ disease and Hashimoto’s thyroiditis). Arch Med Sci [Internet]. 2012;3:415–21. Available from: http://www.termedia.pl/doi/10.5114/aoms.2012.28593.
|
| 22. |
Si X, Zhang X, Tang W, Luo Y. Association between the CTLA-4 +49A/G polymorphism and Graves’ disease: A meta-analysis. Exp Ther Med [Internet]. 2012;4(3):538–44. Available from: https://www.spandidos-publications.com/10.3892/etm.2012.618.
|