Abstract
Background
Ovarian carcinoma, one of the most common gynecological malignancies in Central and Eastern Europe, is characterized by a clinical and genetic heterogeneity with a distinct molecular signature for each histologic subtype.
Materials and methods
Here, we established the frequency of the PIK3CA mutations and amplifications in 100 FFPE tissues with the initial diagnosis of serous ovarian carcinoma. Accordingly, the diagnostic value of combining morphology with genetic and immunohistochemical testing was estimated in this cohort.
Results
The PIK3CA mutations and amplifications were found in 4.1% (4/97) and 7.2% (7/97) of samples, respectively with a higher prevalence in low-grade tumors (p=0.0121). All identified variants were classified as pathogenic missense mutations, located within the PIK3CA mutational hotspots. In the light of the molecular and immunohistochemical results, two tumors with the somatic PIK3CA mutations and strongly positive expression for PI3K and hNF1β were eventually re-classified from serous to clear cell carcinomas after pathological re-evaluation.
Conclusions
These findings demonstrate that the PIK3CA mutational screening facilitated establishing an accurate diagnosis of ovarian carcinomas and, more importantly, might allow for personalized treatment optimization. As the PIK3CA mutations result in the PI3K/AKT pathway deregulation, the individuals with the somatic PIK3CA variants may be eligible for personalized targeted therapies with PI3K inhibitors.
Citation
Żuk M, Koczkowska M, Gorczyński A, Sejda A, Biernat W, Wasąg B. Significance of the PIK3CA mutations in the differential diagnosis of ovarian epithelial carcinoma. Eur J Transl Clin Med. 2018;1(2):23-30Introduction
Besides the tumor suppressor genes BRCA1 (MIM: 113705) and BRCA2 (MIM: 600185) other cancer-related genes have been associated with pathogenesis and progression of ovarian cancer (OC) [1-2]. Indeed, each histological subtype of OC is reflected by the specific tumor molecular signature what enables establishing a widely accepted classification of OC into type I and type II tumors developing through different pathways [3]. Besides the BRCA1/2 susceptibility genes, highgrade serous ovarian tumors (type II) are characterized by genomic instability, including frequent mutations in TP53 (MIM: 191170), NF1 (MIM: 613113) and/or CDK12 (MIM: 615514), while clear cell and endometrioid carcinomas (type I) mostly harbor alterations in the ARID1A (MIM: 603024), PIK3CA (MIM: 171834) or PTEN (MIM: 601728) genes [4].
The PIK3CA gene encoding the catalytic subunit p110α of phosphatidylinositol 3-kinase (PI3K) is mutated or amplified in a broad spectrum of neoplasms including colon, breast, ovarian, endometrial, gastric, thyroid, cervical cancer as well as brain tumors, resulting in the deregulation of the PI3K/AKT signaling pathway [5]. The over-activation of the PI3K/AKT pathway contributes to the progression and tumorigenesis in OC and, importantly, may be followed by resistance to chemotherapy [6-9]. However, recent studies demonstrated that this pathway may play an attractive therapeutic role in OC with several agents, i.e. PI3K inhibitors that are currently being explored in pre- and clinical trials [4,10].
In this study, we established the spectrum and frequency of the PIK3CA mutations and amplifications in 100 formalin-fixed paraffin-embedded (FFPE) tissues with the initial diagnosis of serous OC. In addition, the diagnostic value of combining morphology with genetic and immunohistochemical testing was estimated in the group of individuals with OC.
Materials and Methods
Samples selection and histopathological review
A total of 100 FFPE OC samples were enrolled to the study. All tissue samples were collected between 2008 and 2012 and have been stored in the archives of the Department of Pathomorphology at the Medical University of Gdansk. Hematoxylin-eosin (H&E)- stained tumor tissue sections were reviewed independently by two pathologists (A.G. and W.B.). The initial histological diagnosis of serous OC and the tumor tissue content were evaluated for all samples. To obtain a concentration of at least 50% of cancer cells, tissue macrodissection was performed. Briefly, 84% of individuals (84/100) were diagnosed with high-grade tumors, mostly with IIIB (25%) and IIIC (54%) tumor’s stage. The mean individual age at diagnosis was 60 years (range: 36-81).
The study was approved by the local ethics committee at the Medical University of Gdansk. All individuals provided informed written consent prior to study enrollment.
DNA extraction
Genomic DNA was extracted from the macrodissected FFPE tissues using Cobas DNA Sample Preparation Kit (Roche) following the manufacturer’s protocol. NanoDrop 1000 UV Spectrophotometer and Qubit Fluorometer (ThermoFisher Scientific) were used to check the quantity and quality of isolated DNA.
Mutational analysis
The PIK3CA gene mutation screening of exons 1, 4, 7, 9 and 20 was performed using cobas® PIK3CA mutation kit (Roche) in DNA derived from all FFPE tissue samples following the manufacturer’s protocol. The presence of the pathogenic PIK3CA variants was confirmed by next-generation sequencing (NGS) analysis with Tumor Hotspot MASTR plus kit (Agilent). The somatic origin of the alterations, i.e. the paucity of the mutations in the normal tissues, was determined by bidirectional Sanger sequencing (ABI PRISM 3130, Life Technologies) and by cobas® PIK3CA mutation kit (Roche). In all samples, the comprehensive analysis of the BRCA1/2 genes testing was done as part of our previous study [11].
To explore the presence of the PIK3CA amplification and/or the ploidy, samples were tested by dual in situ hybridization (DISH) method using PIK3CA-specific and centromer 3-specific (CEP3) probes on the BenchMark GX automated system (Ventana Medical Systems, Inc.) according to the manufacturer ’s protocol. A quantitative scoring algorithm was applied to 50 representative non-overlapping nuclei within the invasive region per specimen. To assess gene copy number, PIK3CA and CEP3 signals were counted and used for the resulting ratio calculation. The PIK3CA gene status was classified as non-amplified (PIK3CA/CEP3 ratio <2.0) or amplified (PIK3CA/CEP3 ratio ≥2.0).
The nomenclature of the mutations was based on PIK3CA mRNA sequence NM_006218.2, BRCA1 mRNA sequence NM_007294.3 and BRCA2 mRNA sequence NM_000059.3 according to the recommendations of the Human Genome Variation Society (HGVS).
Immunohistochemical staining
Immunostaining was performed on tissue microarrays (TMAs) of the representative tumor site from FFPE tissue sections. With Manual Tissue Arrayer MTA1 (Beecher Instruments Inc., Sun Prairie, USA), two cylindrical cores of 1.0 mm diameter were removed from each donor paraffin block and transferred to pre-molded recipient paraffin blocks at defined array positions. The presence of PI3K protein in TMAs was assessed by immunohistochemical staining with monoclonal rabbit antibody against p110α PI3K, clone C73F8 (Cell Signaling Technology Inc., Danvers, USA) with dilution 1:50, using the Novolink Polymer Detection System (Novocastra, Wetzlar, Germany) with appropriate positive and negative controls included. Each core was individually evaluated according to the H-score system [12]. To exclude the diagnosis of clear cell carcinoma, all TMAs samples were immunostained with the polyclonal rabbit hNF1- β antibody against human HNF1 homeobox B (Sigma-Aldrich, St. Louis, USA) of 1:100 dilution, and in cases showing any nuclear positivity the staining was repeated on the whole slides
Statistical analysis
For univariate analysis, two-tailed Fisher’s exact test was used to compare categorical variables with a p-value < 0.05 considered as statistically significant. All statistical analyses were performed with GraphPad software.
Results
PIK3CA alterations
The pathogenic PIK3CA mutations and amplifications were identified in 4/97 (4.1%) and 7/97 (7.2%) samples, respectively (Figure 1 and Table 1). Three samples (#8, #34 and #95) were excluded from the molecular analysis due to the low quality of DNA. Among the positive PIK3CA missense variants, c.1634A>C (p. Glu545Ala) and c.1624G>A (p.Glu542Lys) in exon 10 as well as c.3140A>G (p.His1047Arg) and c.3140A>T (p.His1047Leu) in exon 21 were found; each observed in a single tumor as a somatic alteration (samples #22, #44, #20 and #43, respectively). The PIK3CA mutations or amplifications were more common in low-grade tumors compared to high-grade samples (33.3% versus 7.3%; p=0.0121; Table S1). We did not observe the presence of the gene amplification in combination with the PIK3CA point mutation.
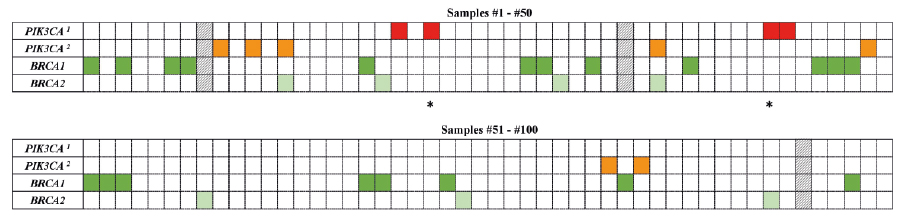
Figure 1. A heat map with the mutation pattern of the PIK3CA and BRCA1/2 genes in ovarian carcinomas
a PIC3CA mutation; b PIC3CA ampflification
The column represents each tumor sample, while each row represents a gene (red and orange –PIK3CA; dark green – BRCA1; light green – BRCA2). Asterisks (*) indicate the tumor samples with diagnosis refinement based on the molecular and immunohistochemical results (#22 and #43). Three samples (#8, #34 and #95), excluded from the study due to the low DNA quality, are shown as the black and white pattern.
Table 1: Spectrum of the PIK3CA alterations in ovarian carcinomas
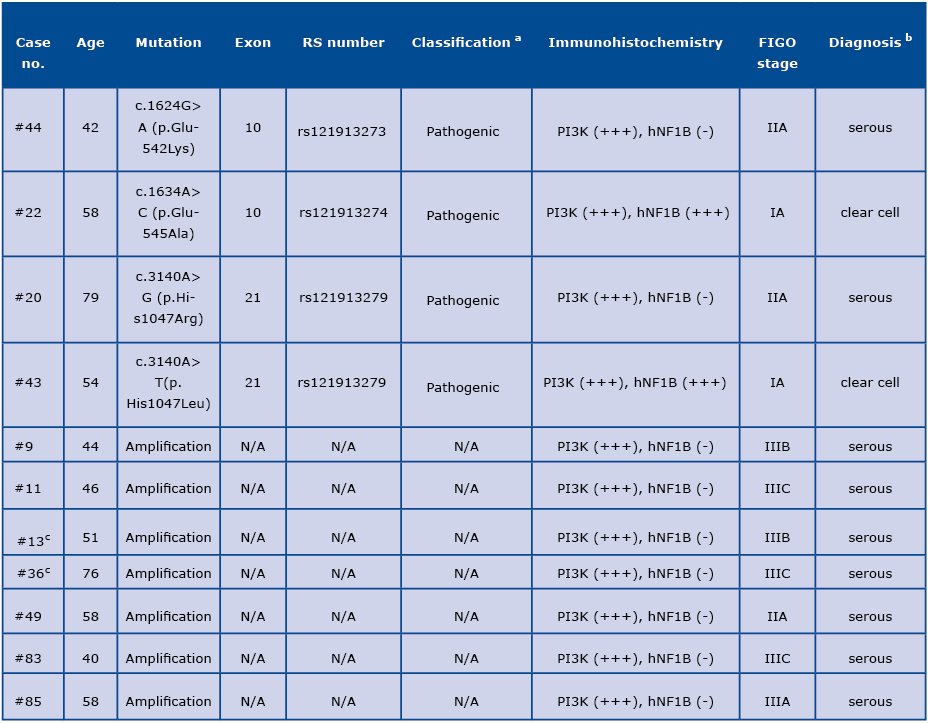
a Variants’ classification reported in the publicly available databases, including ClinVar and HGMD.
b Based on the molecular and immunohistochemical analysis, in two cases (#22 and #43) the initial diagnosis of serous ovarian carcino
ma was refined to the clear cell ovarian carcinoma (in italics) after histological re-evaluation by two independent pathologists.
c Tumors with presence of an additional pathogenic variant in the BRCA2 gene (#13 and #36).
N/A: not applicable
Pathogenic variants in BRCA1/2
Of the 11 tumors with the PIK3CA mutation or amplification, two samples (#13 and #36) had an additional variant in the BRCA2 gene: c.5328dupT (p.Leu1777Glufs*4) and c.5993_5997dupAAGTG (p.Phe2000Lysfs*6), respectively (as detailed in Table S2). Due to the lack of normal tissue for case subject #13, the germline origin of the aforementioned alteration was confirmed only in sample #36. The comprehensive BRCA1/2 analysis in the studied material was performed by NGS as part of our previous study [11].
Immunohistochemical analysis and diagnosis refinement
Based on the routine histopathological examination, initially all samples were classified as serous OC (Figure 2A). Additional immunohistochemical analysis revealed a high expression (+++) for PI3K in all tumors carrying the PIK3CA mutations or amplifications (Figure 2C-D). Moreover, two samples (#22 and #43) were strongly positive (+++) for hNF1β (Figure 2E). In the light of immunohistochemical and genetic results, these samples were eventually diagnosed as clear cell carcinomas after independent re-evaluation by two expert pathologists. The remaining tumors were characterized by negative (-) or weak (+) PI3K and hNF1β expression (Figure 2F). The algorithm of the study approach is shown in Figure 3.
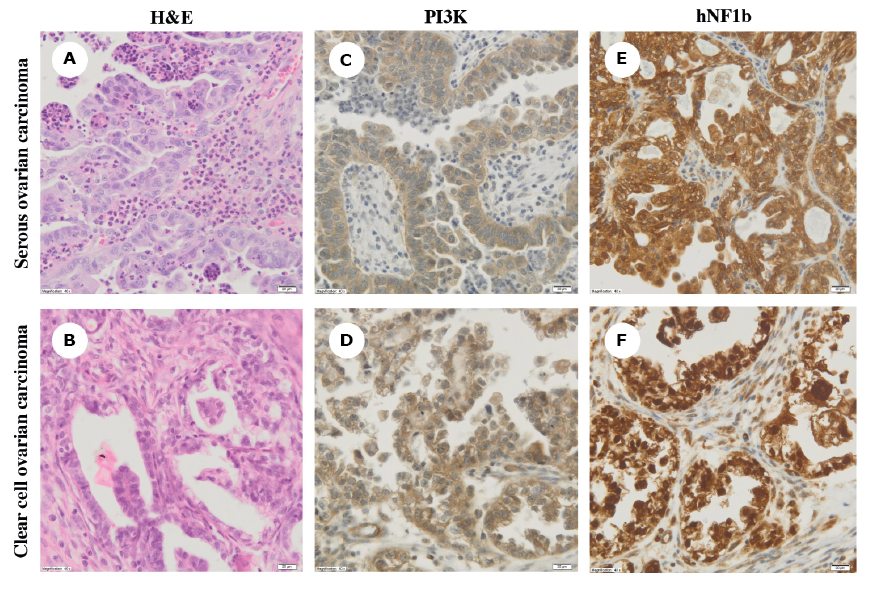
Figure 2 A-F. Hematoxylin and eosin (H&E) staining (panels A-B) and immunohistochemical analysis of phosphatidylinositol 3-kinase (PI3K) and hNF1b expression (panels C-D and E-F, respectively) in serous and clear cell ovarian carcinomas (magnification, ×40)
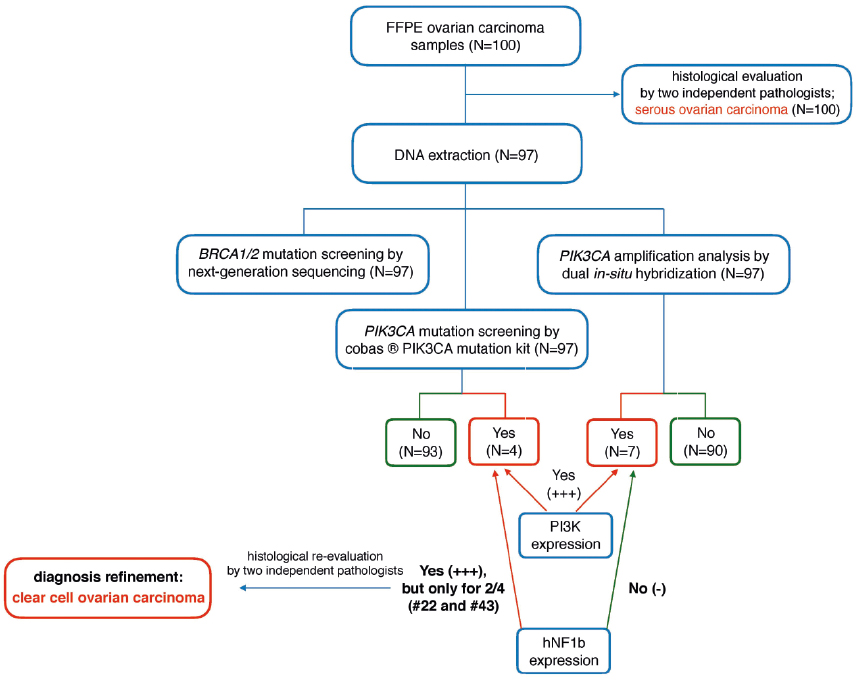
Figure 3. Algorithm of the study approach in the differential diagnosis of epithelial ovarian carcinomas
Discussion
As OC are characterized by a clinical and genetic heterogeneity with different treatment sensitivity and prognosis, it is critical to make a definitive histological diagnosis. In the current study, the spectrum and frequency of the PIK3CA mutations and amplifications in 100 FFPE samples with the initial diagnosis of serous OC was established. Additionally, we estimated the efficacy of implementation the expanded genetic testing panel, comprising PIK3CA mutation screening, into the routine diagnostic procedures in OC.
The spectrum of the PIK3CA mutations has been widely studied in numerous neoplasms, including breast and ovarian cancer [5]. Although PIK3CA is one of the most commonly mutated oncogenes in human malignancies with over 13,000 tissue samples carrying the PIK3CA pathogenic variants (as reported in the COSMIC database as of November 2018), the prevalence of the somatic PIK3CA mutations in OC is moderate. As the overall frequency is estimated at 4-12%, depending on the histological subtype, the finding of the PIK3CA variants in 4.1% (4/97) of the samples in ourcohort is in line with expectations [13-16]. Campbell et al. (2005) and Wang et al. (2005) reported that the PIK3CA variants predominantly clustered among clear cell and endometrioid OC compared to serous carcinomas (20% versus 2.3%), suggesting different mechanisms of their pathogenesis [14,16]. In this study, the presence of the PIK3CA mutations was observed in tumors with the initial diagnosis of serous carcinomas that may likely explain such a low detection rate. Moreover, two PIK3CA mutational hotspots locations have been described in the highly conserved regions within the helical and kinase domains of the PI3K subunits (exon 10 and 21, respectively) with the estimated frequency of over 80% of all somatic missense mutations [5]. Functional studies demonstrated that these mutations affect the cellular transformation in vivo and in vitro by increasing the kinase activity [17-18]. In this study, the PIK3CA variants were identified exclusively within these hotspots, suggesting functional importance.
The PIK3CA amplification, another mechanism activating PI3K/AKT pathway, has been reported in OC more frequently than point mutations (16%-25% versus 4-12%), but no clear bias towards any particular histological subtype was observed [14,19-21]. Taken together, the overall frequency of the PIK3CA mutations and amplifications in the studied cohort was 11.3% (11/97) that is slightly lower compared to previous reports [14,21]. Indeed, the PIK3/AKT pathway is reported to be activated in approximately one third of OC cohort, but comprising mostly of samples with the clear cell histological subtype [4]. In this study, all tumors represented serous OC, mostly high-grade (84% with the stage III), except for two cases with the diagnosis refinement to clear cell OC (Table 1 and Figure 3). That likely explains the differences in the overall frequency of the PIK3CA alterations between our studied group and the previously described cohorts.
With the rapid development and accessibility of genomic technology, genetic testing is becoming increasingly important in the clinical management of cancer patients and expecting to be widely implemented in routine oncological practice. That facilitates not only establishing an accurate diagnosis, but, more importantly, allows for personalized treatment optimization. As demonstrated by Italiano et al. (2016), the molecular genetic results enabled for the diagnosis refinement in ~14% (53/384) of soft tissue sarcomas samples, suggesting that genetic testing should be a mandatory step in soft tissue sarcomas diagnostic approach even if the histological diagnosis is made by the expert pathologists in the field [22]. In this study, two tumors (#22 and #43) were wrongly classified as serous OC prior to molecular testing (Figure 3). Only the identification of the PIK3CA mutations, commonly present in endometrioid and clear cell OC, casted a shadow of doubt on the accuracy of the initial histological diagnosis in these samples [3-4]. Therefore, we believe that genomic profiling of tumors, including at least mutational screening of the cancer-related genes specific for each histological subtype (i.e. PIK3CA, PTEN, ARID1A, NF1 and/or CDK12) might offer benefits also in the differentiation of OC.
Although the BRCA1/2 mutation-positive tumors represent the most common group of OC tumors and these women are eligible for the poly(ADP-ribose) polymerase 1 inhibitors (iPARP1) targeted therapy [23], there is a need to develop new effective treatment alternatives. Another advantage of the PIK3CA mutation screening in OC is the possibility of the application the targeted therapies in individuals with the PIK3CA mutation-positive tumors. As the PI3K/AKT pathway is frequently deregulated in selected histological subtypes of OC, inhibition of PI3K seems to be a worthwhile treatment strategy in these individuals. To date, several PI3K inhibitors have been tested in pre- and clinical trials and the most promising results are observed in combination with other targeted therapies, such as MEK inhibitors, anti-angiogenic therapy and hormonal therapy in selected OC individuals [24-25]. Moreover, the PIK3CA alterations are reported to play a significant role in acquiring chemoresistance in OC cells, all the more so, it is critical to diagnose those tumors early enough to implement an appropriate treatment in these individuals and consequently improve their clinical prognosis [7-8,21].
As effective cancer treatment starts with the correct pathological diagnosis, genetic testing may play a complementary role in this critical diagnostic process. In the current study, the initial misdiagnosis of two OC samples was re-evaluated after providing the additional molecular and immunohistochemical results. Moreover, we confirmed the frequency of the PIK3CA mutations and amplifications in OC samples, and more importantly, demonstrated that the genomic profiling along with the detailed histological and immunohistochemical examinations may resolve the diagnostic dilemma. Although the PIK3CA alterations affect only a small percentage of OC tumors, it is still important to evaluate their mutational status because these individuals may be eligible for the personalized targeted therapies.
Abbreviations
OC: ovarian carcinoma; FFPE: formalin-fixed paraffin-embedded tissue sample; PI3K: phosphatidylinositol 3-kinase; NGS: next-generation sequencing; H&E: hematoxylin & eosin; DISH: dual in situ hybridization; TMA: tissue microarray
Acknowledgements
We thank all the individuals who agreed to participate in the study.
Online databases
ClinVar: https://www.ncbi.nlm.nih.gov/
Cosmic: http://cancer.sanger.ac.uk/cosmic
GraphPad: https://www.graphpad.com/
HGMD: http://www.hgmd.cf.ac.uk/ac/index.php
HGVS: http://varnomen.hgvs.org/
OMIM: https://www.omim.org/
References
| 1. |
Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science (80- ). 1994;266(5182):66-71.
|
| 2. |
Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378(6559):789-92.
|
| 3. |
Kurman RJ, Shih I-M. The dualistic model of ovarian carcinogenesis: revisited, revised, and expanded. Am J Pathol. 2016;186(4):733–47.
|
| 4. |
Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. Lancet. 2014;384(9951):1376-88.
|
| 5. |
Samuels Y, Waldman T. Oncogenic Mutations of PIK3CA in Human Cancers BT - Phosphoinositide 3-kinase in Health and Disease: Volume 2. Rommel C, Vanhaesebroeck B, Vogt PK, editors. Phosphoinositide 3-kinase Heal Dis Vol 2 Curr Top Microbiol Immunol. 2010;347:21-41.
|
| 6. |
Dobbin ZC, Landen CN. The importance of the PI3K/AKT/MTOR pathway in the progression of ovarian cancer. Int J Mol Sci. 2013;14(4):8213-27.
|
| 7. |
Hu L, Hofmann J, Lu Y, Mills GB, Jaffe RB. Inhibition of phosphatidylinositol 3′-kinase increases efficacy of paclitaxel in in vitro and in vivo ovarian cancer models. Cancer Res. 2002;62(4):1087-92.
|
| 8. |
Ohta T, Ohmichi M, Hayasaka T, Mabuchi S, Saitoh M, Kawagoe J, et al. Inhibition of phosphatidylinositol 3-kinase increases efficacy of cisplatin in in vivo ovarian cancer models. Endocrinology. 2006;147(4):1761-9.
|
| 9. |
Lee S, Choi E-J, Jin C, Kim D-H. Activation of PI3K/Akt pathway by PTEN reduction and PIK3CA mRNA amplification contributes to cisplatin resistance in an ovarian cancer cell line. Gynecol Oncol. 2005;97(1):26-34.
|
| 10. |
Banerjee S, Kaye SB. New strategies in the treatment of ovarian cancer: current clinical perspectives and future potential. Clin cancer Res. 2013;19:961-8.
|
| 11. |
Koczkowska M, Zuk M, Gorczynski A, Ratajska M, Lewandowska M, Biernat W, et al. Detection of somatic BRCA 1/2 mutations in ovarian cancer–next‐generation sequencing analysis of 100 cases. Cancer Med. 2016;5(7):1640-6.
|
| 12. |
Pirker R, Pereira JR, von Pawel J, Krzakowski M, Ramlau R, Park K, et al. EGFR expression as a predictor of survival for first-line chemotherapy plus cetuximab in patients with advanced non-small-cell lung cancer: analysis of data from the phase 3 FLEX study. Lancet Oncol. 2012;13(1):33-42.
|
| 13. |
Levine DA, Bogomolniy F, Yee CJ, Lash A, Barakat RR, Borgen PI, et al. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin cancer Res. 2005;11(8):2875-8.
|
| 14. |
Campbell IG, Russell SE, Choong DYH, Montgomery KG, Ciavarella ML, Hooi CSF, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64(21):7678-81.
|
| 15. |
Campbell IG, Russell SE, Phillips WA. PIK3CA mutations in ovarian cancer. Clin Cancer Res. 2005;11(19):7042-3.
|
| 16. |
Wang Y, Helland Å, Holm R, Kristensen GB, Børresen‐Dale A. PIK3CA mutations in advanced ovarian carcinomas. Hum Mutat. 2005;25(3):322.
|
| 17. |
Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci. 2006;103(5):1475-9.
|
| 18. |
Ikenoue T, Kanai F, Hikiba Y, Obata T, Tanaka Y, Imamura J, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65(11):4562-7.
|
| 19. |
Willner J, Wurz K, Allison KH, Galic V, Garcia RL, Goff BA, et al. Alternate molecular genetic pathways in ovarian carcinomas of common histological types. Hum Pathol. 2007;38(4):607-13.
|
| 20. |
Woenckhaus J, Steger K, Sturm K, Münstedt K, Franke FE, Fenic I. Prognostic value of PIK3CA and phosphorylated AKT expression in ovarian cancer. Virchows Arch. 2007;450(4):387-95.
|
| 21. |
Kolasa IK, Rembiszewska A, Felisiak A, Ziolkowska-Seta I, Murawska M, Moes J, et al. PIK3CA amplification associates with resistance to chemotherapy in ovarian cancer patients. Cancer Biol Ther. 2009;8(1):21-6.
|
| 22. |
Italiano A, Di Mauro I, Rapp J, Pierron G, Auger N, Alberti L, et al. Clinical effect of molecular methods in sarcoma diagnosis (GENSARC): a prospective, multicentre, observational study. Lancet Oncol. 2016;17(4):532-8.
|
| 23. |
Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, et al. Oral poly (ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376(9737):245-51.
|
| 24. |
Gasparri ML, Bardhi E, Ruscito I, Papadia A, Farooqi AA, Marchetti C, et al. PI3K/AKT/mTOR pathway in ovarian cancer treatment: are we on the right track? Geburtshilfe Frauenheilkd. 2017;77(10):1095-103.
|
| 25. |
Cheaib B, Auguste A, Leary A. The PI3K/Akt/mTOR pathway in ovarian cancer: therapeutic opportunities and challenges. Chin J Cancer. 2015;34(1):4-16.
|